I am trying over-plot some empirical data with error bars on top of my modelled data. The error bars seem to be rendering first and are consequently getting over written (see below)
I have tried using zorder but I still get the same result. The code I am using is
for i in range(1,len(pf)):
pf[i,:] = av_pf_scale * pf[i,:]
pylab.semilogy(pf[0,0:180],pf[i,0:180],color='0.75')
pylab.semilogy(av_pf[0:180],color='r')
pylab.semilogy(av_mie[0:180],color='g', linestyle='-')
pylab.draw()
f = pylab.errorbar(ang,data[j],
yerr = delta_data[j],
fmt = 'o',
markersize = 3,
color = 'b',
zorder = 300,
antialiased = True)
I would appreciate if anyone can tell me how to make the errorbars render on top.


 Hacky work around:
Hacky work around: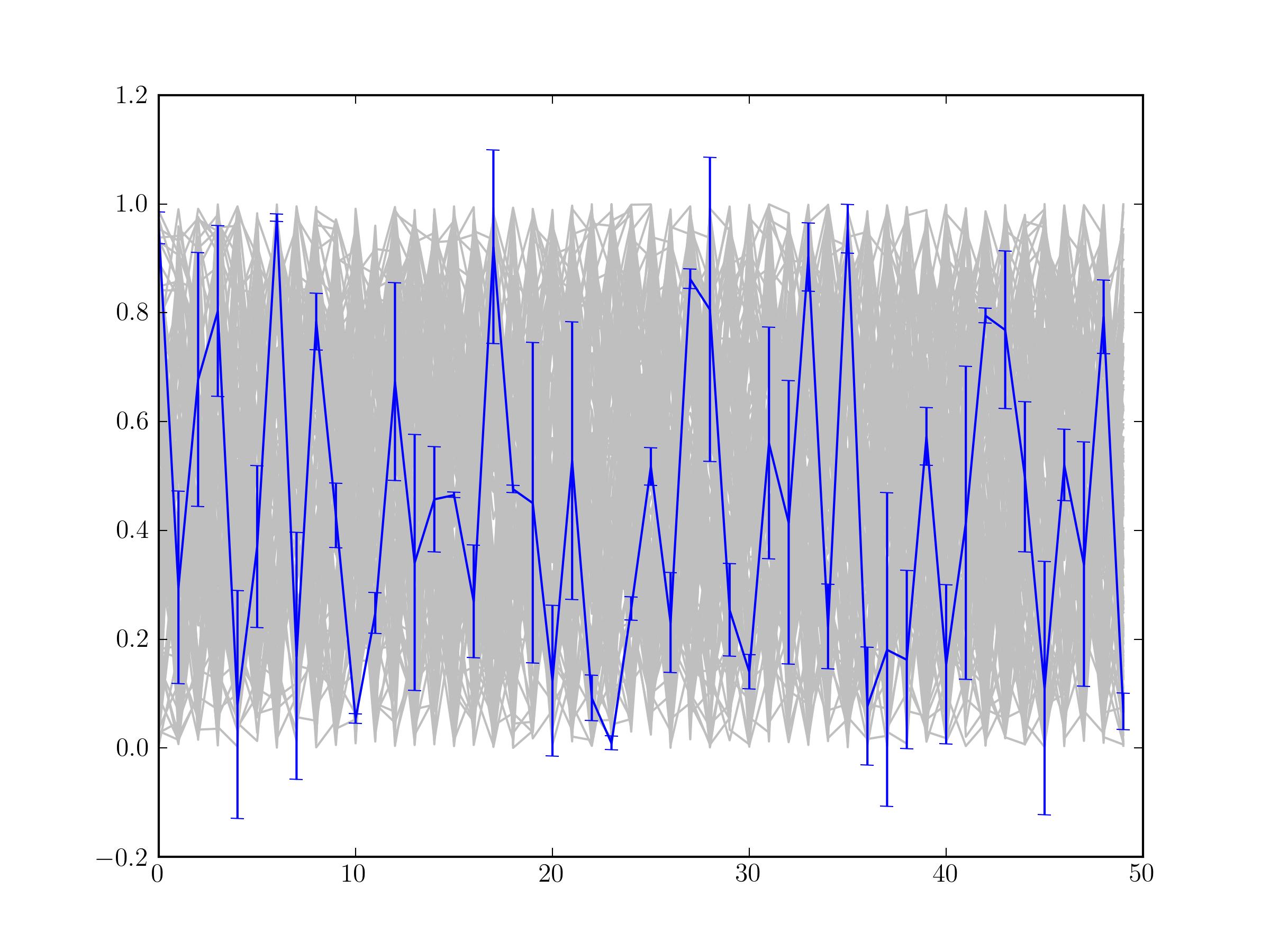





 已为社区贡献126445条内容
已为社区贡献126445条内容

所有评论(0)