Currently seaborn offers functionality for split violinplots by setting split=True, according to a hue variable. I would like to make a 'half' violin plot, i.e. a plot where half of each violin is omitted. Such a plot depicts something similar to a pdf for each continuous variable, plotted on one side of each vertical line of each categorical variable only.
I have managed to trick seaborn to plot this with an extra data point outside the plotted range of values and an extra dummy hue, but I would like to know if this can be done without actually altering the dataset, e.g. within sns.violinplot() arguments.
For instance, this graph:
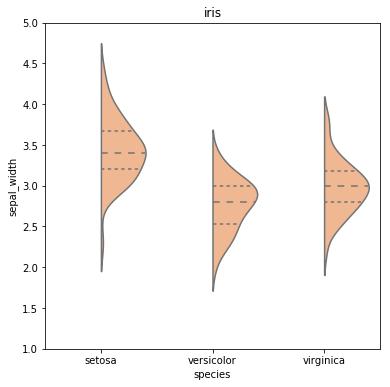
Was created by this snippet:
# imports
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# load dataset from seaborn
datalist = sns.get_dataset_names()
dataset_name = 'iris'
if dataset_name in datalist:
df = sns.load_dataset(dataset_name)
else:
print("Dataset with name: " + dataset_name + " was not found in the available datasets online by seaborn.")
# prepare data
df2 = df.append([-999,-999,-999,-999,'setosa'])
df2['huecol'] = 0.0
df2['huecol'].iloc[-1]= -999
# plot
fig = plt.figure(figsize=(6,6))
sns.violinplot(x='species',y="sepal_width",
split=True, hue ='huecol', inner = 'quartile',
palette="pastel", data=df2, legend=False)
plt.title('iris')
# remove hue legend
leg = plt.gca().legend()
leg.remove()
plt.ylim([1,5.0])
plt.show()
I was looking for a solution similar to this but did not find anything satisfactory. I ended up calling seaborn.kdeplot multiple times as violinplot is essentially a one-sided kernel density plot.
Example
Function definition for categorical_kde_plot below
categorical_kde_plot(
df,
variable="tip",
category="day",
category_order=["Thur", "Fri", "Sat", "Sun"],
horizontal=False,
)
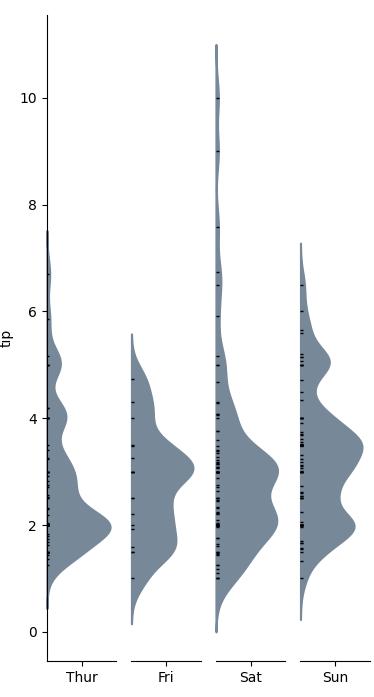
with horizontal=True, the output would look like:
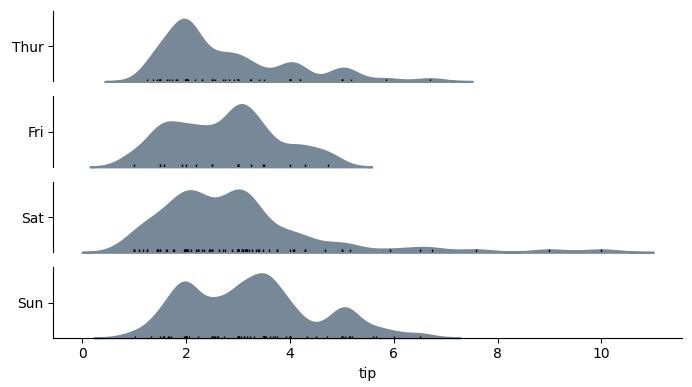
Code
import seaborn as sns
from matplotlib import pyplot as plt
def categorical_kde_plot(
df,
variable,
category,
category_order=None,
horizontal=False,
rug=True,
figsize=None,
):
"""Draw a categorical KDE plot
Parameters
----------
df: pd.DataFrame
The data to plot
variable: str
The column in the `df` to plot (continuous variable)
category: str
The column in the `df` to use for grouping (categorical variable)
horizontal: bool
If True, draw density plots horizontally. Otherwise, draw them
vertically.
rug: bool
If True, add also a sns.rugplot.
figsize: tuple or None
If None, use default figsize of (7, 1*len(categories))
If tuple, use that figsize. Given to plt.subplots as an argument.
"""
if category_order is None:
categories = list(df[category].unique())
else:
categories = category_order[:]
figsize = (7, 1.0 * len(categories))
fig, axes = plt.subplots(
nrows=len(categories) if horizontal else 1,
ncols=1 if horizontal else len(categories),
figsize=figsize[::-1] if not horizontal else figsize,
sharex=horizontal,
sharey=not horizontal,
)
for i, (cat, ax) in enumerate(zip(categories, axes)):
sns.kdeplot(
data=df[df[category] == cat],
x=variable if horizontal else None,
y=None if horizontal else variable,
# kde kwargs
bw_adjust=0.5,
clip_on=False,
fill=True,
alpha=1,
linewidth=1.5,
ax=ax,
color="lightslategray",
)
keep_variable_axis = (i == len(fig.axes) - 1) if horizontal else (i == 0)
if rug:
sns.rugplot(
data=df[df[category] == cat],
x=variable if horizontal else None,
y=None if horizontal else variable,
ax=ax,
color="black",
height=0.025 if keep_variable_axis else 0.04,
)
_format_axis(
ax,
cat,
horizontal,
keep_variable_axis=keep_variable_axis,
)
plt.tight_layout()
plt.show()
def _format_axis(ax, category, horizontal=False, keep_variable_axis=True):
# Remove the axis lines
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
if horizontal:
ax.set_ylabel(None)
lim = ax.get_ylim()
ax.set_yticks([(lim[0] + lim[1]) / 2])
ax.set_yticklabels([category])
if not keep_variable_axis:
ax.get_xaxis().set_visible(False)
ax.spines["bottom"].set_visible(False)
else:
ax.set_xlabel(None)
lim = ax.get_xlim()
ax.set_xticks([(lim[0] + lim[1]) / 2])
ax.set_xticklabels([category])
if not keep_variable_axis:
ax.get_yaxis().set_visible(False)
ax.spines["left"].set_visible(False)
if __name__ == "__main__":
df = sns.load_dataset("tips")
categorical_kde_plot(
df,
variable="tip",
category="day",
category_order=["Thur", "Fri", "Sat", "Sun"],
horizontal=True,
)









 已为社区贡献126445条内容
已为社区贡献126445条内容

所有评论(0)